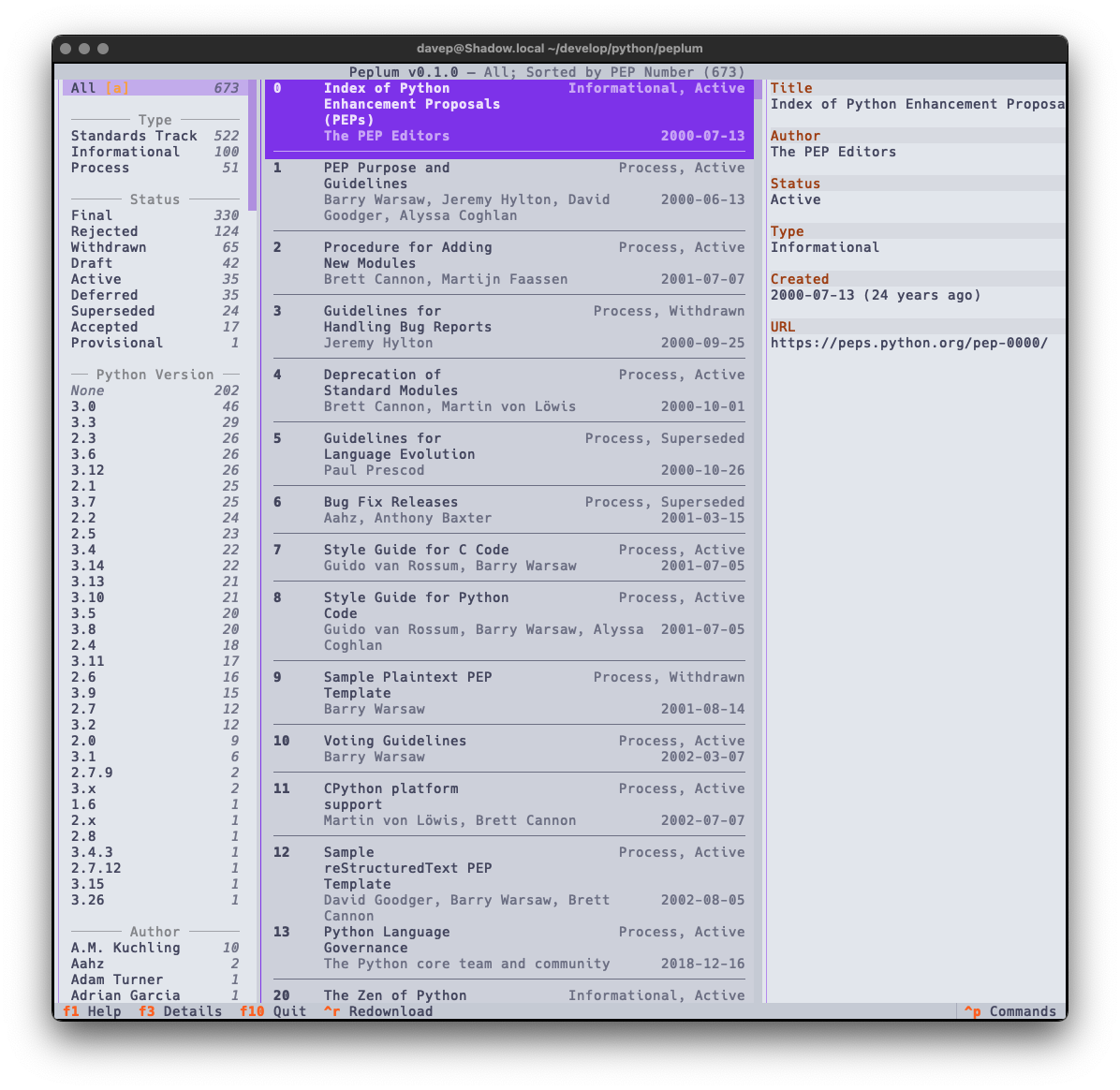
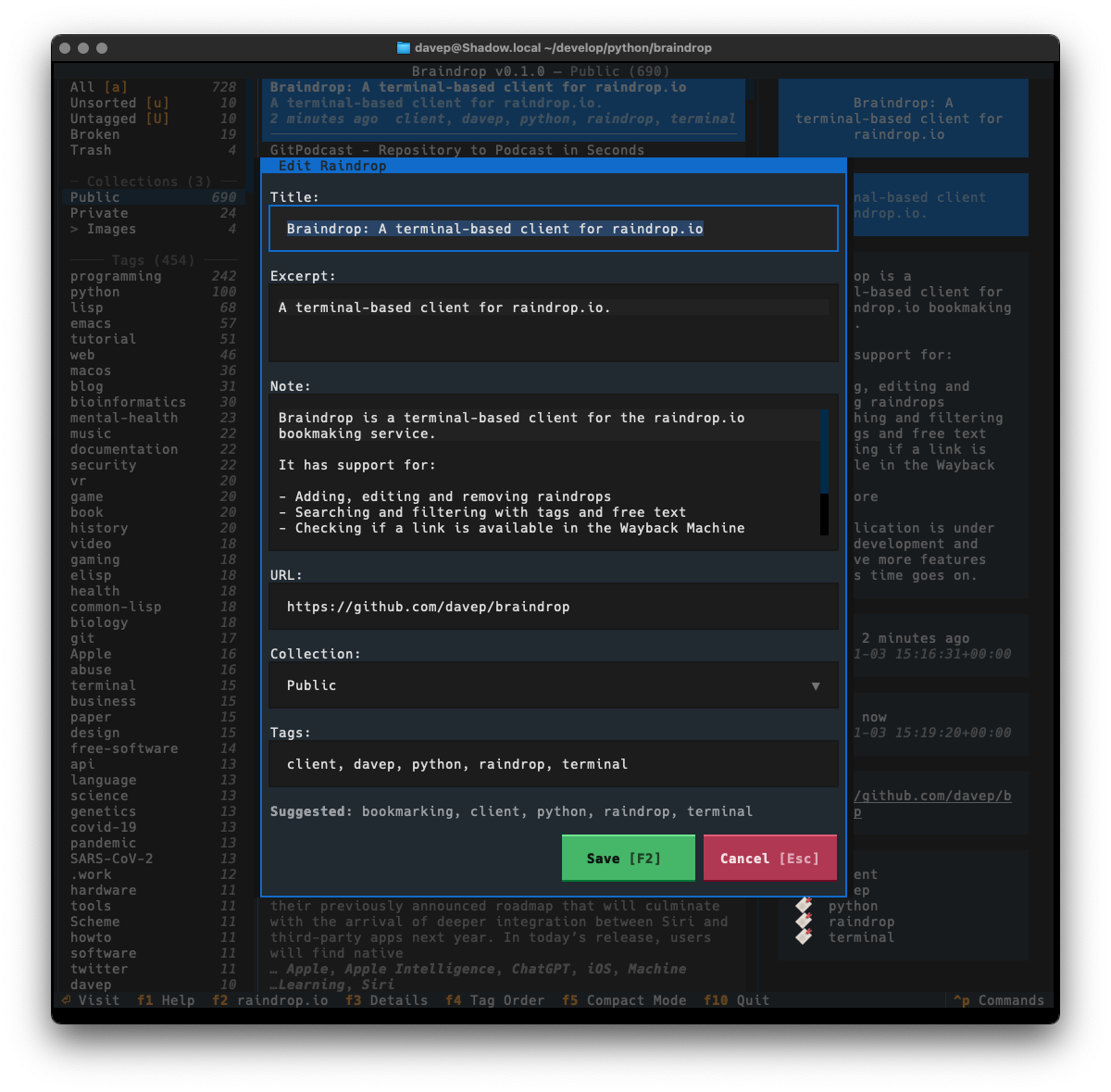
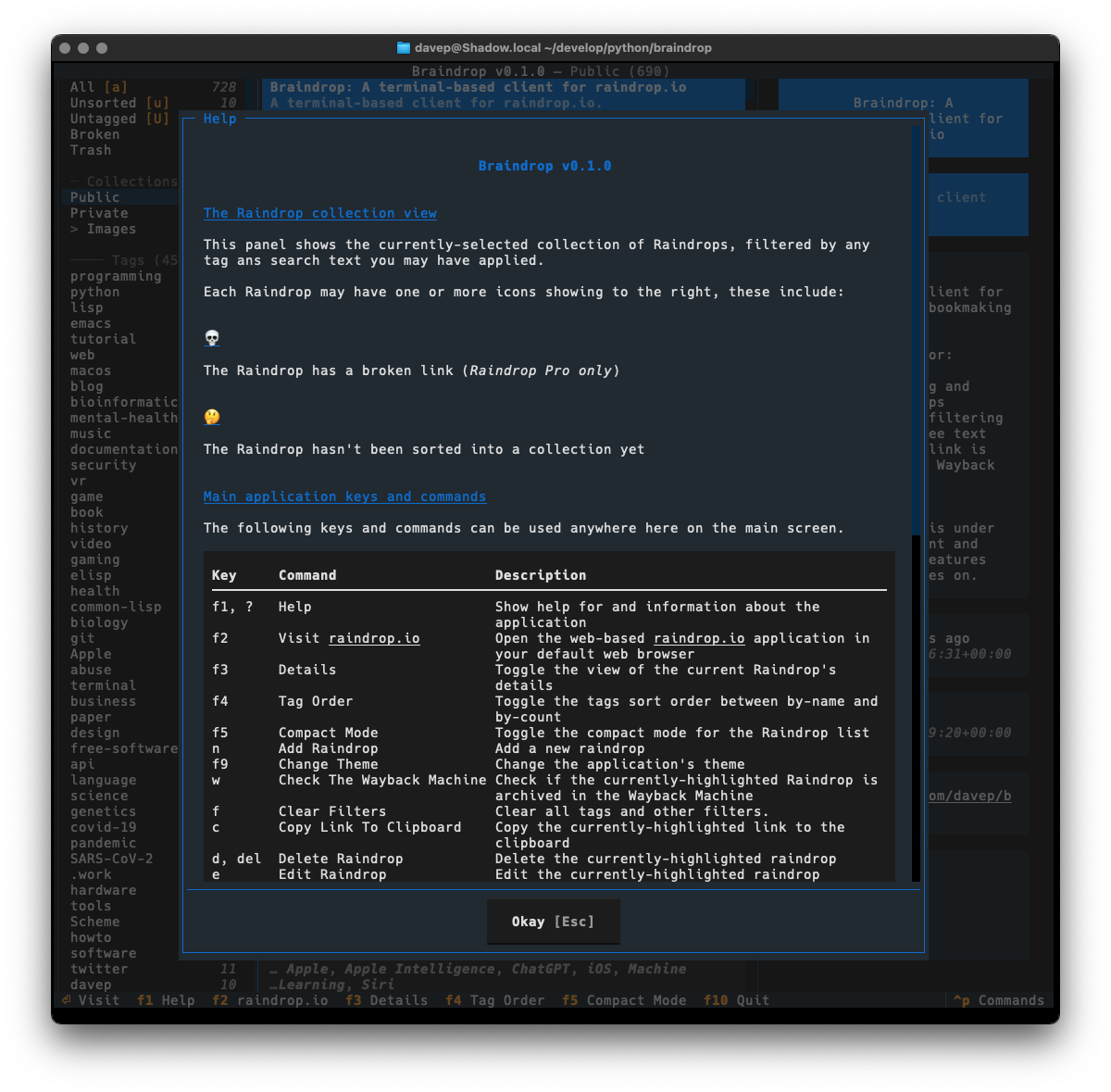
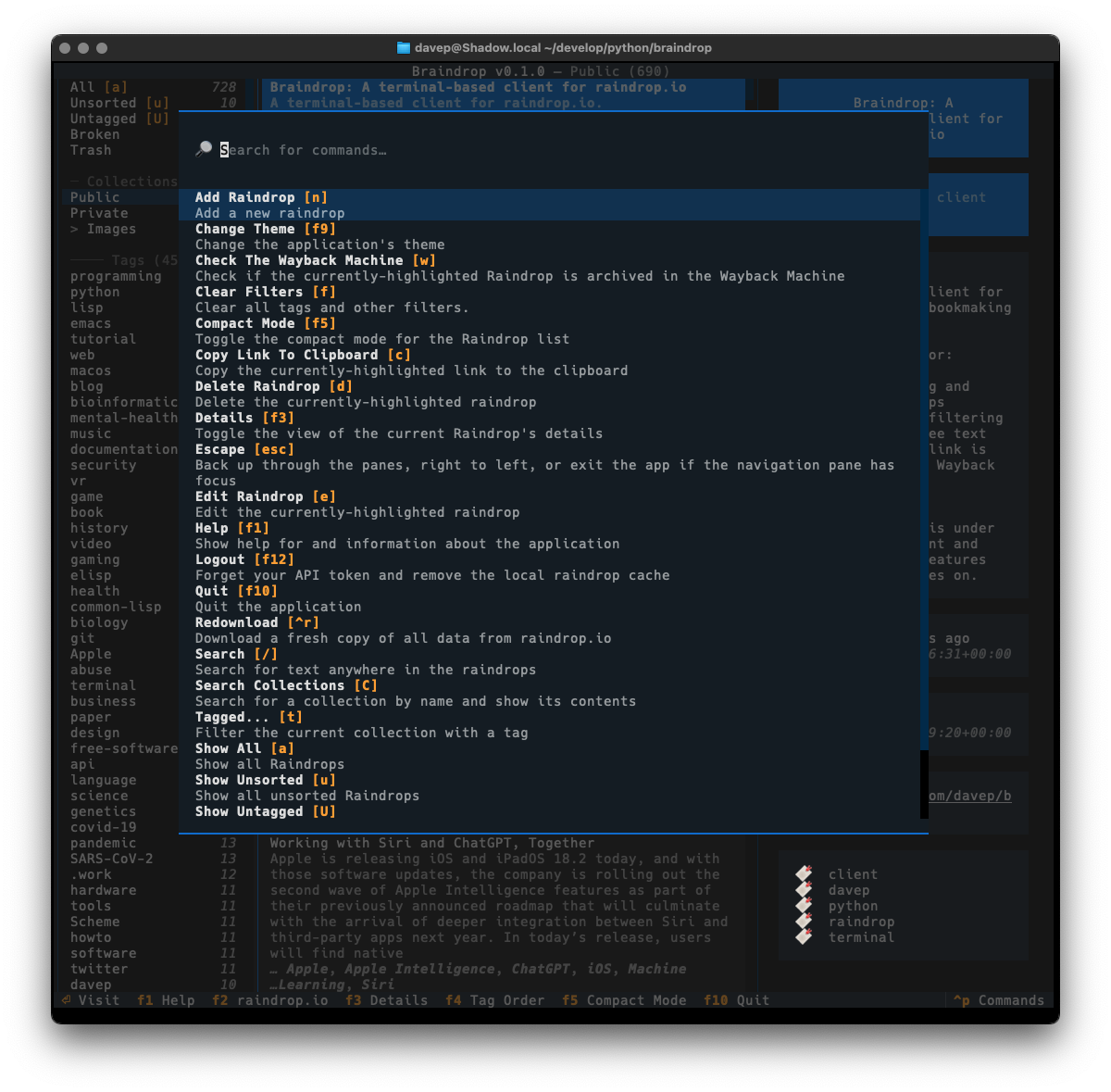
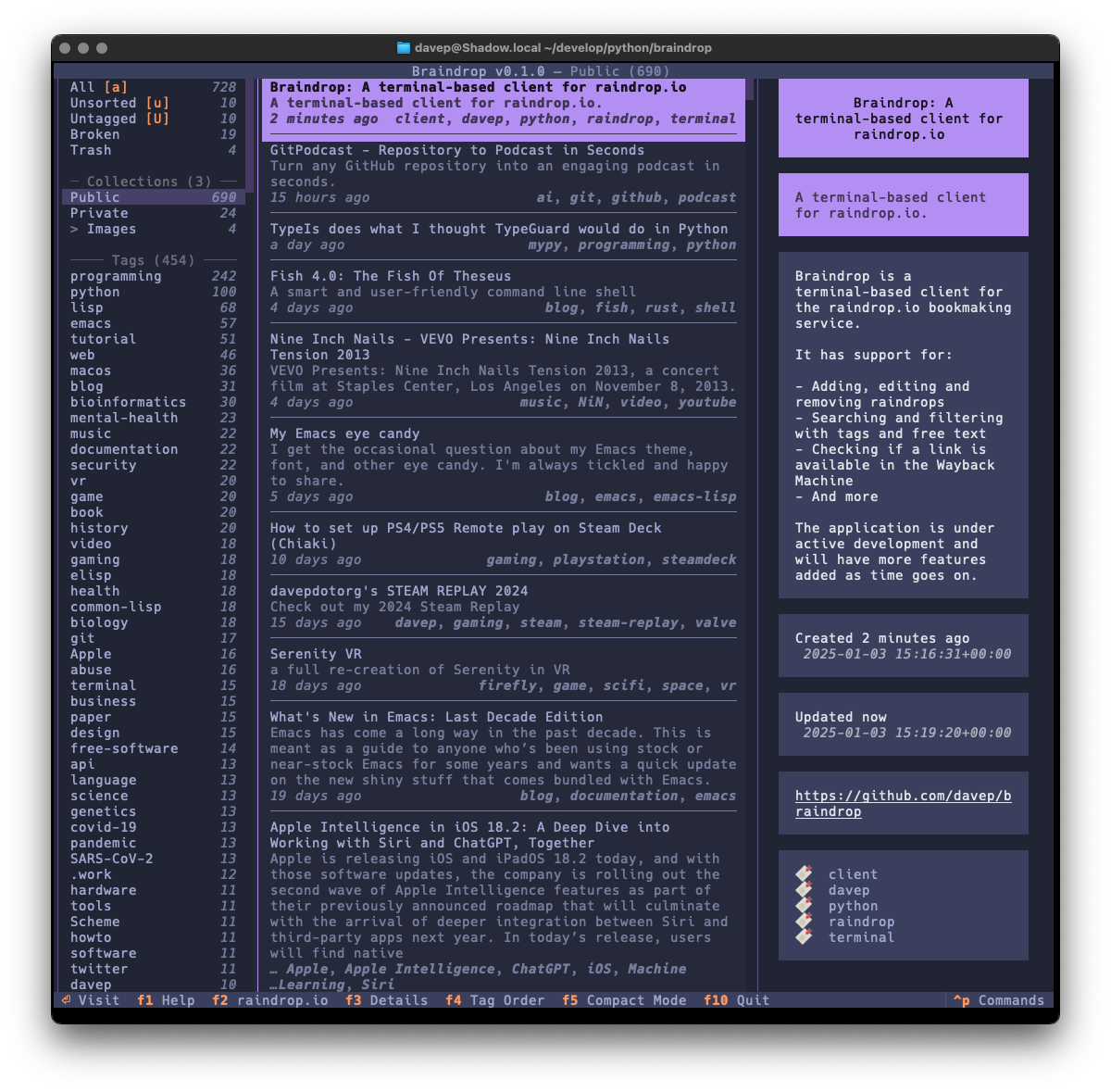
Recently I've been on a bit of a "turn stuff into Markdown files and slap
them in an Obsidian Vault" trip. This kicked off a couple of months back
when I made a decision unrelated to coding.
On and off, since my teenage years, I've kept journals. Since those teenage
years it's been more off than on, but a couple of times in my adult life
it's been really helpful to actually write one. The last time this happened
was early 2019. It was pretty vital I did that at the time and it was a
really sensible and helpful decision, and an approach to the situation I was
in that I'd recommend to anyone (and have done on occasion to anyone going
through the same thing).
The actual motivation for starting that particular journal is long behind
me, but I'd got into the habit of writing it and so, until a couple or so
months back, I kept jotting something down every day. But I came to the
realisation that I didn't need to and that it had become something of a
chore.
I'd been using an application called Journey. It's
a great app, does the job well, but was also suffering from the creep of
"AI" (I've had a few apps ion my arsenal that don't need it, acquire a
useless "AI" feature). This privacy-problematic change of direction,
combined with the realisation that I didn't need to write about my day,
every day, any more, made me decide it was time to stop and cancel the
subscription.
Thankfully Journey has a pretty comprehensive export option so I used it and
didn't think too much more about it for a while.
Meanwhile I also had a subscription to Evernote that
I didn't really use any more. Within it I had held a handful of years of
journal entries from a decade or so ago, along with other "remember this
for some point in the future" stuff. For the longest time I was on some
really cheap tier that didn't exist any more, one that was low enough that I
didn't really notice the cost go out each month so I kept putting off
exporting things and closing it all down until "next month".
Then I got an email from them to say they were forcing me onto some new tier
that was more expensive. So that was the final straw there. I made an export
of what I had in Evernote and closed that account down too.
A wee while went past and then I got to thinking that it might be
interesting to try and combine both these sources into one archived journal.
I had stuff from around 2010 to 2015, and I also had stuff from 2019 until
2024; the former in the Evernote archive and the latter in the Journey
archive. Surely I could write a couple of tools to turn that data into one
consolidated Obsidian Vault?
Over the course of a couple of weekends
journey2md and
evernote2md were born. While both
of those tools work differently, they're both designed to populate the same
Obsidian Vault. Once I was happy with this I did the mass conversion and I
was happy with the result.
Now I have years of journal entries, all converted to Markdown files and
made available for reading via an application that lets me rummage through
history using dates and tags and all sorts of other searching.
So I was happy with that and didn't give it much more though.
Then last week I got to thinking...
Twitter has turned into the worst place possible and I can't for the life of
me think why any right-thinking person who has an ounce of humanity or has
anything approaching a humanistic outlook on life would remain an active
user. Honestly I stuck it out longer than was sensible, but in June 2023 I
finally quit for good.
Back when the new owner was
confirmed
I, like a lot of people, extracted my archive. It's since been sat in
storage doing nothing, yet there's a lot of data in there that could be
interesting to work with, or just to go back and look through. So last
week's thought was "why don't I also turn this into an Obsidian Vault?".
So I did...
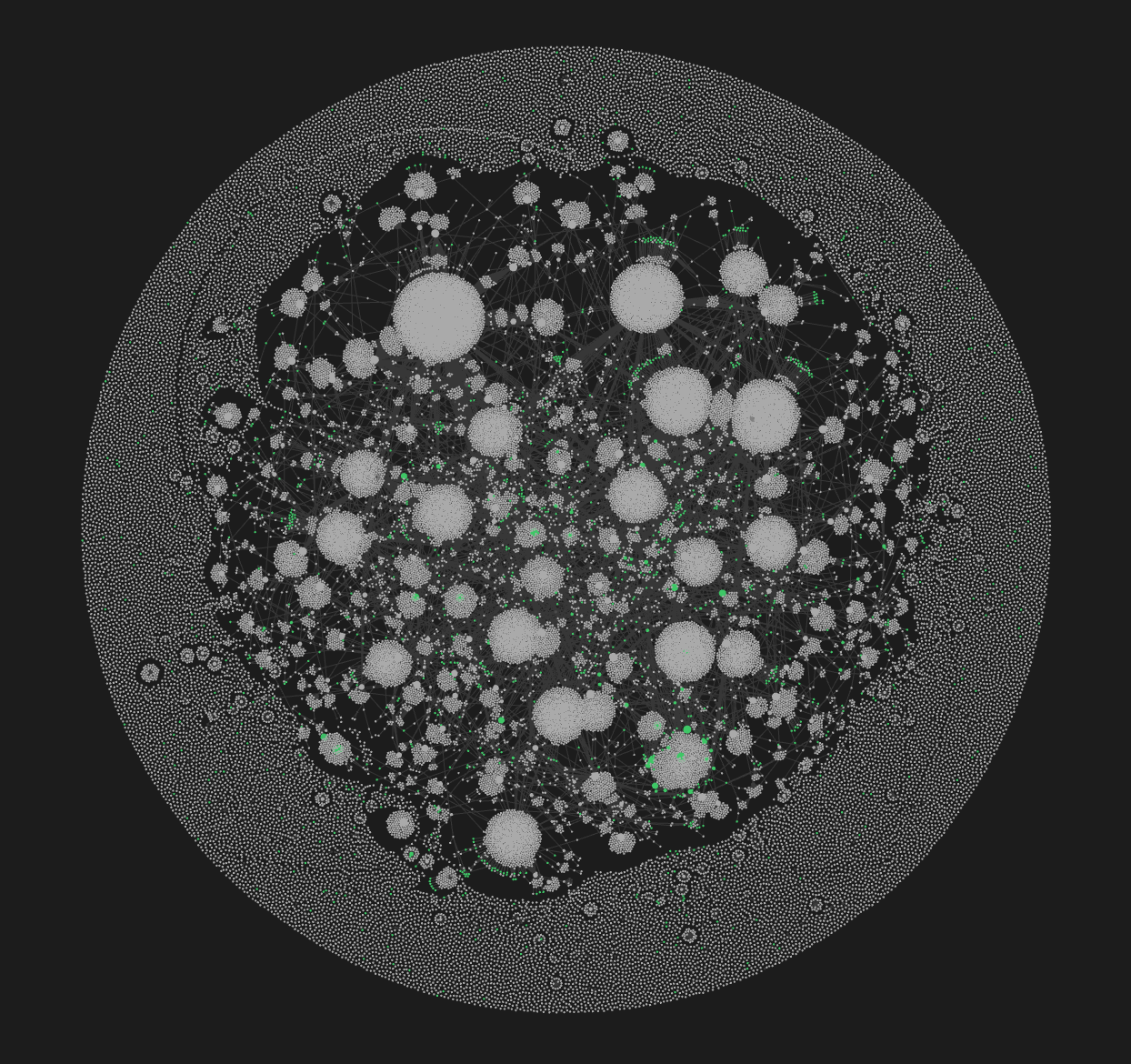
The tool I built to do this is
bird2glass. As you'll see in the
README it makes a
few assumptions about the state of Twitter archive dumps and also what a
user wants from this. Personally I'm pleased with the result.
The main aim of the tool is to break the tweets down into a hierarchy of
year, month and day...
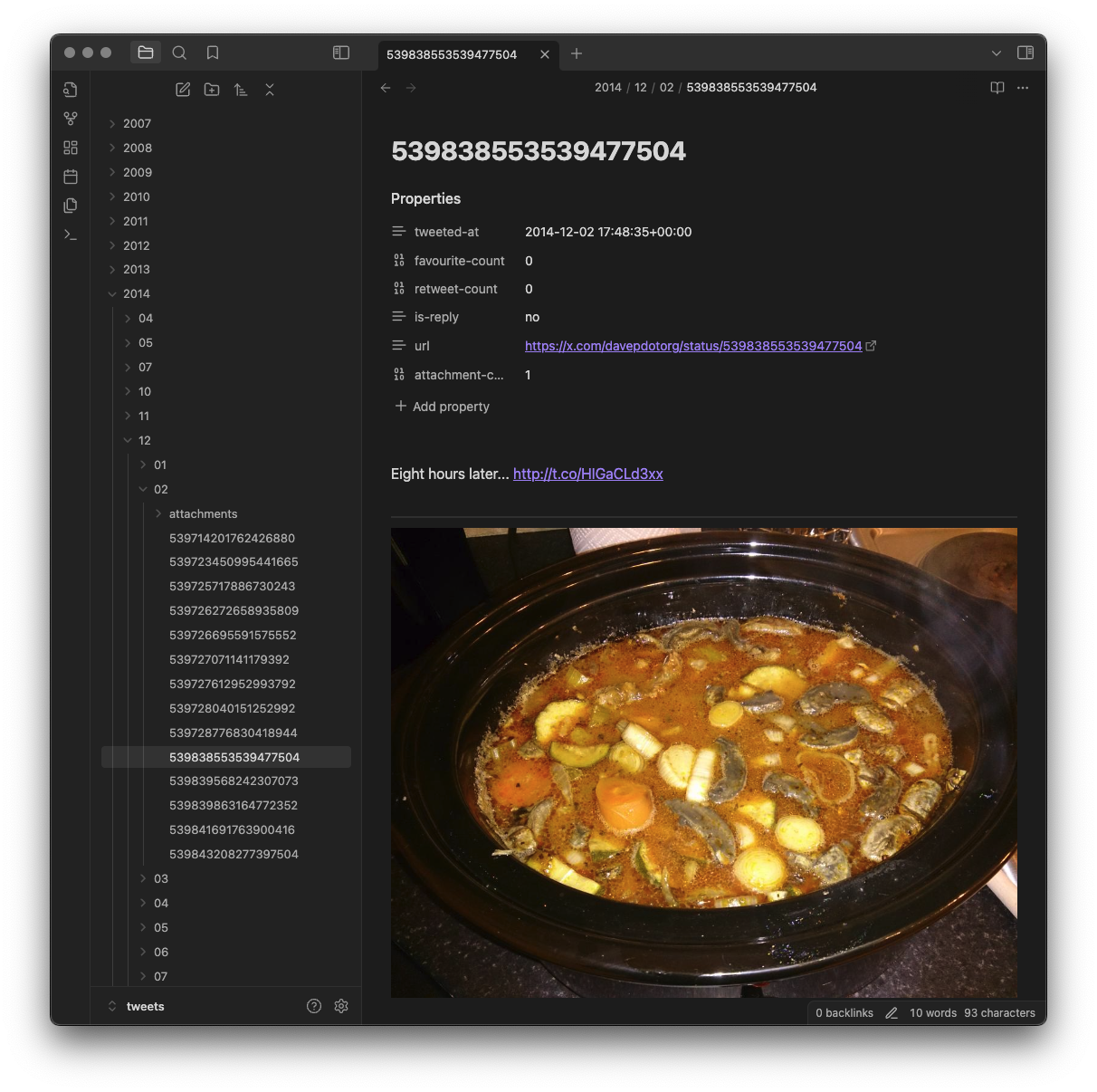
...and also to connect them with any account that was being replied to or
mentioned in some way...
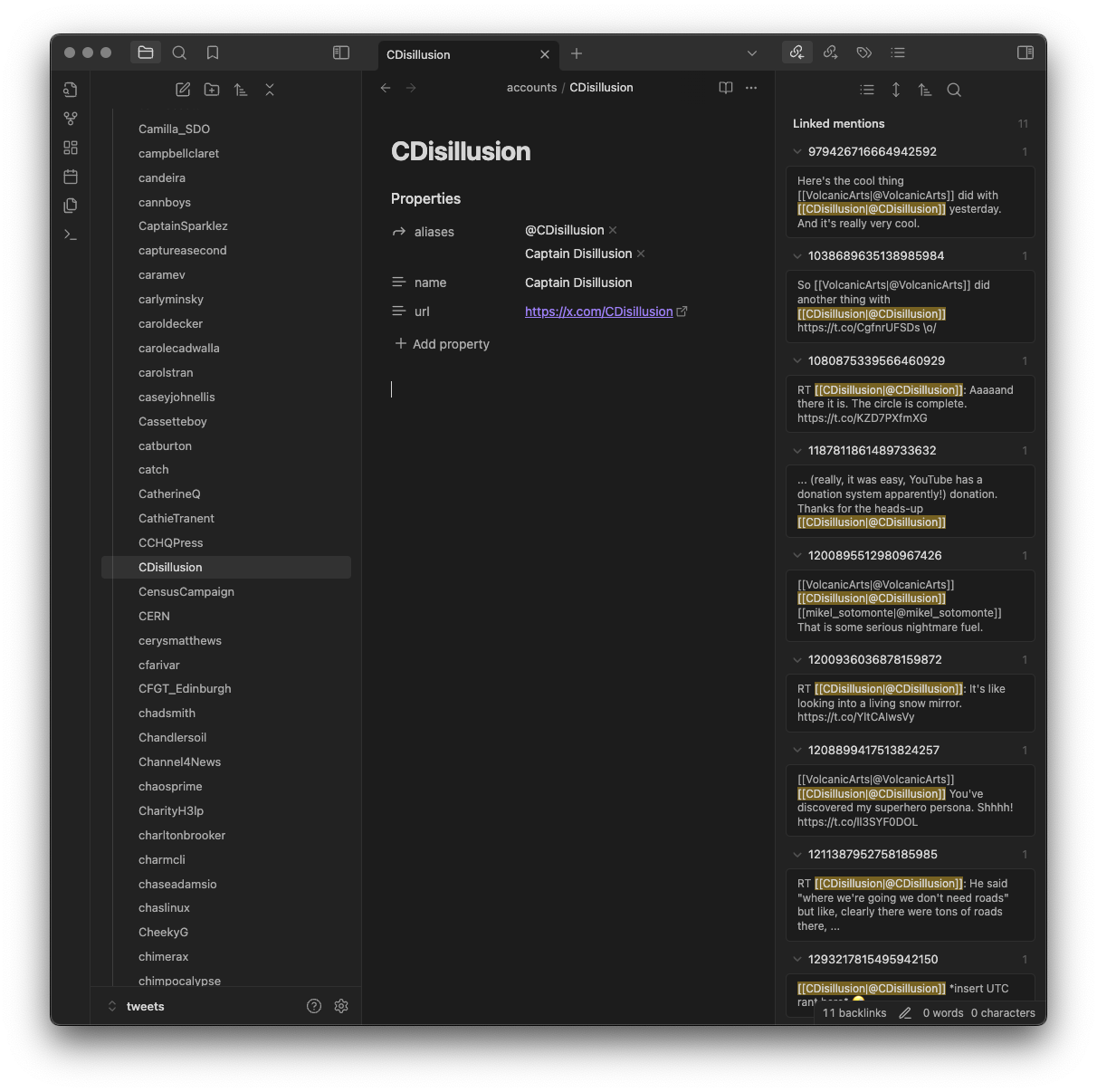
This user view is handy when viewing backlinks, as it gives you a list of
all the tweets that mention that user (and, of course, if you're into
Obsidian's graph it will make for some interesting connections within
there).
I sense there's more I can do with this, and I imagine I will continue to
tinker with it. Meanwhile though, if that sounds like something you'd
benefit from do feel free to grab it and play with it and hack on it. Keep
in mind the notes and assumptions that are in the README, and really be
prepared for a lot of files to be created if you did a lot of tweeting like
I did (I do think that over 50,000 individual files for an Obsidian Vault is
a bit silly, if I'm honest).
Meanwhile... I might need to look at other applications and think about how
I can turn the data into useful Markdown collections!